I think it is important to ground ourselves in some of the computational concepts that are common among data scientists and bioinformaticians. This can again help to remove the black box of the computer and the terminal, and can improve our productivity as scientists. With that, I want to step back and just sort of reintroduce the concept of commands from the command line from a conceptual standpoint.
What is a command? A command is basically just a program or an app. It is a chunk of code that someone has written that takes
input, processes that input, and produces output. The really common and useful chunks of code (e.g. ls
or cd) have become mainstays in modern operating systems to the extent that we don't even need to
think about the underlying code, but it is there.
The other main difference between commands and modern programs or apps is that they are invoked almost entirely in a text-based terminal program. This provides versatility, as commands can be scripted to run in sequence, as well as the power to convey complex instructions to the computer.
Imagine being in a country where you don't know the language. Maybe you go into a coffee shop and want to order something, but without knowledge of the language, you basically have to resort to pointing at what you want on the menu. This is really effective! It can convey a lot of meaning and be really quick to get across some ideas. But there may be details that you can't convey because they aren't on the menu, like that you wanted extra sugar in your coffee.
But if you knew the language you could easily get this information and much more complex and nuanced information across in your order. This is the difference between using the mouse to convey instructions to the computer, and using text-based commands, and opens up a wide-range of possibilities for the user of the computer... though just like learning a new spoken language, it can be difficult to grasp at first.
Most commands, and most data science in general, relies on plain-text formatted documents and input. When it comes down to it, a command is really just:
I refer to this as the central dogma of data science.
This means 2 things:
- Text formatting is very important. Knowing the expected input format for a command means you can format your data correctly
- Being able to easily view and manipulate text files becomes crucial for a productive data scientist
We'll learn some common bioinformatics data formats in this workshop and throughout ConGen, but for now let's focus on point 2: viewing and manipulating text with text editors
There are many programs you could use as a text editor, ranging from the most basic text editing capabilities, to full blown integrated development environments (IDEs) with syntax highlighting for different languages, remote syncing, git integration and so on.
The RStudio interface we're using right now is an example of an IDE, centered around the R programming language, and contains it's own text editor.
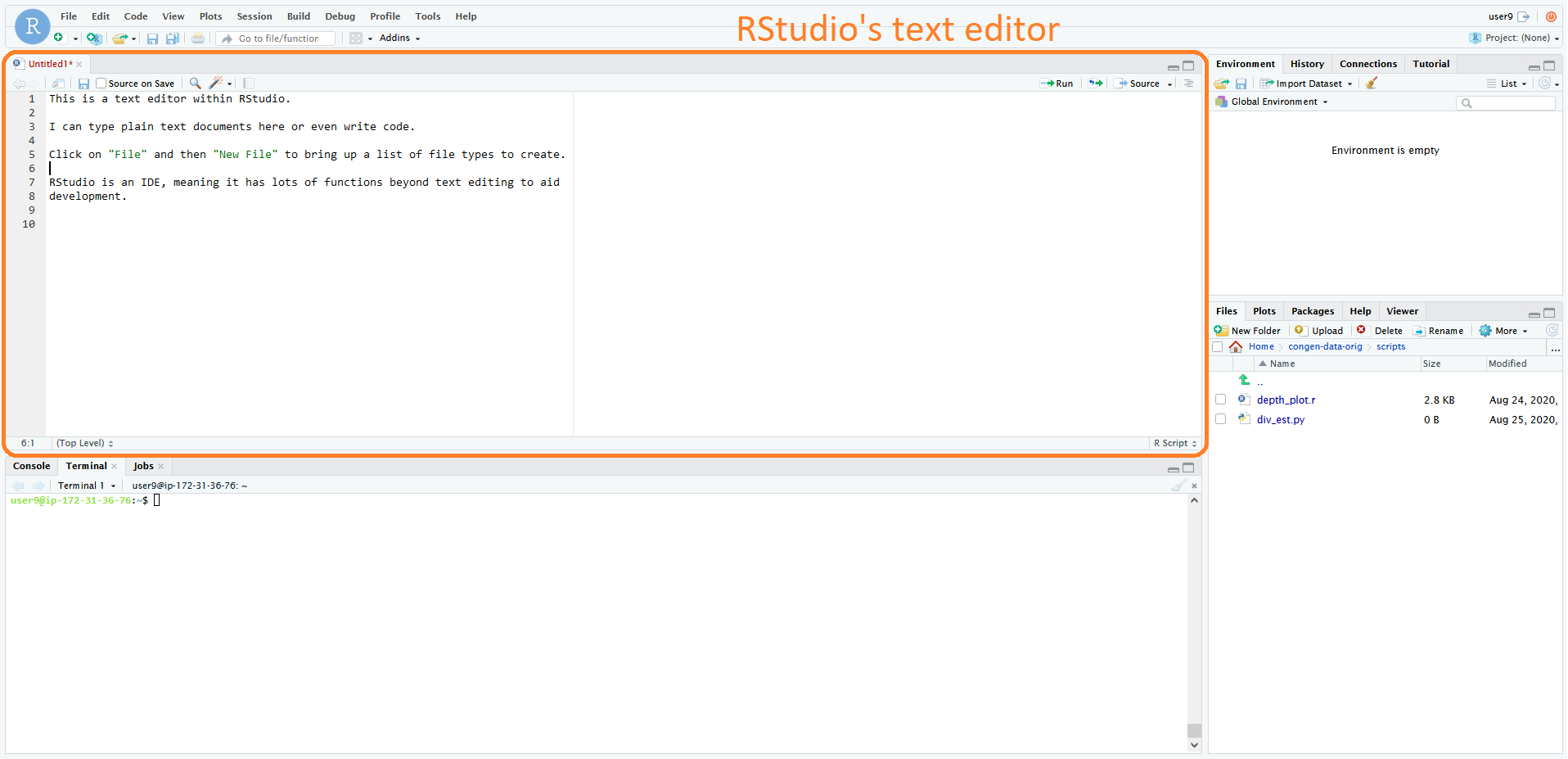
Text editors are also available within Linux terminals themselves, in the form of programs such as nano,
Emacs, or vim. See this table for a list of some
text editors. You should find one that suits your style to make you a more productive bioinformatician.
After years of trying many of the options listed in the table, I have settled on VScode as my text editor. It provides many of the capabilities of an IDE, but keeps true to it's basic function: editing text. The remote addon has also been a gamechanger for editing files on remote servers.
The last conceptual topic I want to discuss is the philosophy for command development, which leads nicely into our activities.
In the late 1970's as the Unix operating system was being developed, the software engineers settled on a set of norms for the programs they wrote. Their philosophy basically boiled down to two things, for our purposes:
- Write a program that does one thing and does that thing really well
- Write programs that can work with each other -- the output of one program should become the input of another.
This is opposed to other software design philosophies, for instance Microsoft, which designs software to do one thing well, but also gives that software to ability to do numerous other things, maybe not optimally. For example, the primary purpose of Microsoft Word is word processing -- writing and formatting text. But within work you can also make tables, edit images, and a plethora of other things that have little to do directly with word processing.
ls; top) and
a Microsoft program (Word).
This does not mean that Unix commands lack versatility. They often have input options that you can read about using the
man (manual) command. For instance, by typing man ls, you will bring up
a screen that shows all of ls's options. The main thing to notice is that these options all work towards the main goal of the
ls program: listing directory contents to the screen.
You can read more about the Unix philosophy here.
The second point of the Unix philosophy leads us to a couple of powerful ways for manipulating text from the command line: piping and redirecting text output from a command.
Piping output from one command to another is done with the vertical bar character (|). By default,
most Unix commands print their output directly to the screen. What the pipe says is that, instead of printing the output to the screen,
use that output as the input of another command. For instance, we know that ls lists the contents of a
directory by printing them to the screen. But what if we're searching for a specific file? Well we could pipe the output from
ls to our string search program, grep:
ls | grep "interesting-filename"| Command line parameter | Description |
|---|---|
| ls | The Linux list directory contents command. With no path specified, this lists the contents of the current directory. |
| | | The pipe character, telling the shell to use the output from the previous command as the input to the next command. |
| grep | A Linux string search and pattern matching command that takes as input a file or stream from a pipe and searches for a given string. |
| "interesting-filename" | The string we want grep to search for in the given input. |
Now we would expect only files with the string "interesting-filename" in them to be printed to the screen, because we've piped our
ls output to grep.
Redirecting command output is another essential command line function, and is accomplished with the greater than
character (>). Redirecting takes the output that would have been printed to the screen and instead
writes it to a specified file. For instance, if we want to save contents of our current directory to a file, we could run:
ls > dir-contents.txt| Command line parameter | Description |
|---|---|
| ls | The Linux list directory contents command. With no path specified, this lists the contents of the current directory. |
| > | The redirect character, telling the shell to save the output from the previous command to the specified file. |
| dir-contents.txt | The file we want to save the output to. |
Now, if we were to open the file dir-contents.txt in our favorite text editor, we would see
the list of files saved there.
Piping and redirecting can be used in conjunction with each other:
ls | grep "interesting-filename" > interesting-dir-contents.txt| Command line parameter | Description |
|---|---|
| ls | The Linux list directory contents command. With no path specified, this lists the contents of the current directory. |
| | | The pipe character, telling the shell to use the output from the previous command as the input to the next command. |
| grep | A Linux string search and pattern matching command that takes as input a file or stream from a pipe and searches for a given string. |
| "interesting-filename" | The string we want grep to search for in the given input. |
| > | The redirect character, telling the shell to save the output from the previous command to the specified file. |
| interesting-dir-contents.txt | The file we want to save the output to. |
Now we've taken the output from ls, piped it to grep to search for filenames
containing a string, and redirected that output to a file called interesting-dir-contents.txt. If we were
to open this file we'd see only the files in the current directory that have the string "interesting-file" in their name.
Also of note, you are not limited in the number of pipes you can do. This means that you could start with text input in a file and process it with several commands via piping to drastically change the information that is output. It is an extremely powerful method of scripting!
You should be aware that if you redirect with > to an existing file that the contents of that
file will be overwritten without warning!
If you have a file you want to redirect output to without overwriting, use two redirect characters in order to append to the file:
ls >> dir-contents.txtNow that we have the conceptual basics of commands reviewed, we'll put these in practice by analyzing some biological data from the command line!

