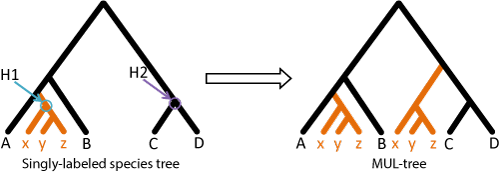
GRAMPA: Gene-tree Reconciliation Algorithm with MUL-trees for Polyploid Analysis
GRAMPA uses gene tree topologies for inferring the presence and mode of polyploidy in a set of species, placing a whole genome duplication event on a phylogeny, and counting gene duplications and losses in the presence of polyploidy.
Install
conda install -c bioconda grampaOtherwise, just download the program straight from github by clicking the download button below. The program is just a python script with no other dependencies, so it should work out of the box!
Citation
Thomas GWC, Ather SH, Hahn MW. 2017. Gene tree reconciliation with MUL-trees to resolve polyploid analysis. Systematic Biology. 66(6):1007-1018. 10.1093/sysbio/syx044





