1. Run info (phyloacc-post)
The PhyloAcc post-processing script was run on Sunday Oct 23, 2022 at 11:31:54 EDT on bioinf02.rc.fas.harvard.edu as follows:
/n/home07/gthomas/projects/PhyloAcc/src/PhyloAcc-interface/phyloacc_post.py -i st-test-branches/ -bf1 4 -bf2 4 -bf3 4
Batch summary
| Complete ST batches | 82 |
| Complete GT batches | 0 |
| Incomplete ST batches | 0 |
| Incomplete GT batches | 0 |
| Loci per batch | 25 |
| Threads per batch | 8 |
| Average runtime per batch (min.) | 50 |
Bayes factor cut-offs
| Bayes Factor | Models compared | Cut-off | # loci above cut-off |
|---|---|---|---|
| BF1 | M1 (target acceleration) vs. M0 (no acceleration) | 4.0 | 1679 |
| BF2 | M1 (target acceleration) vs. M2 (free acceleration) | 4.0 | 818 |
| BF3 | M2 (free acceleration) vs. M0 (no acceleration) | 4.0 | 1140 |
Result summary
Acclerated loci (targets) are those above the cut-off of both BF1 and BF2.
| Total loci | 2029 |
| Accelerated loci (targets) | 784 |
| Accelerated loci (full) | 595 |
See the full log file for more info.
Below are some summary plots. Raw data is also available in in the results folder.
2. Bayes factors
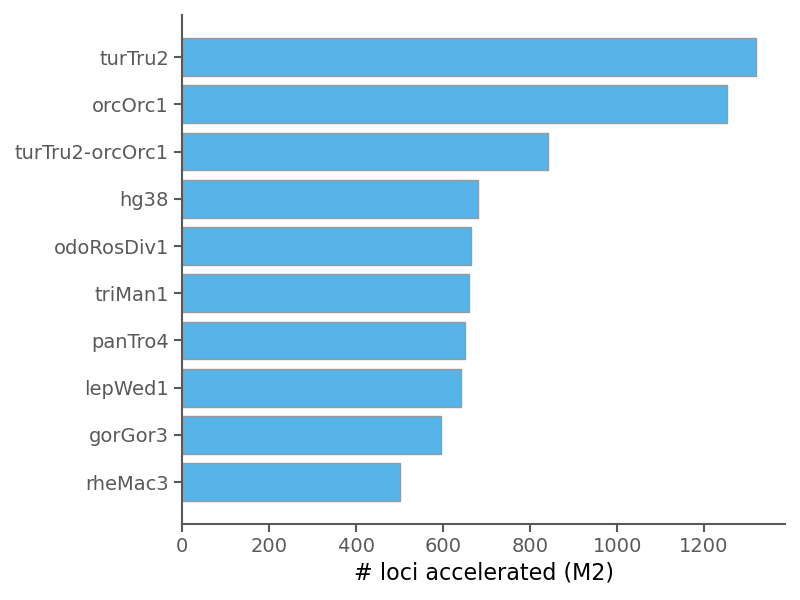
| Median BF1 | 7.854 |
| Median BF2 | 3.754 |
| Median BF3 | 4.882 |
3. Accelerated lineages per locus under full model (M2)
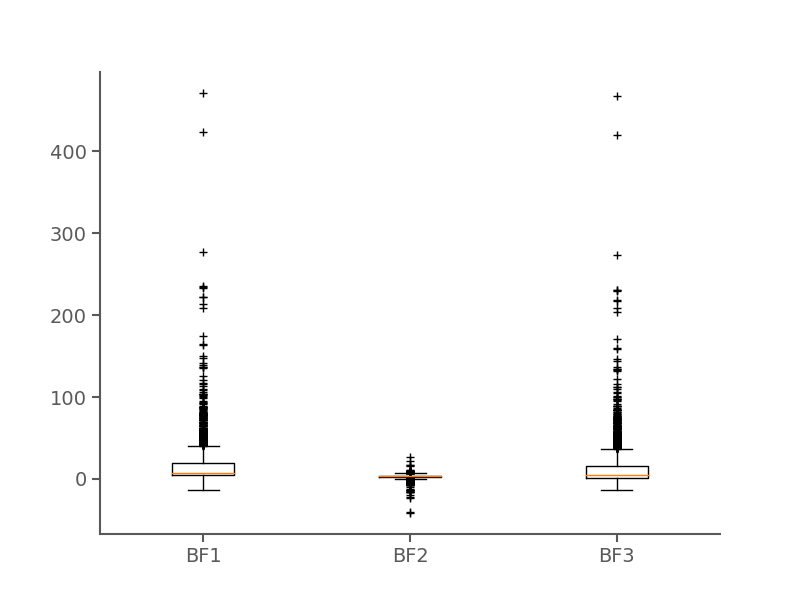
| Avg. accelerated branches per locus | 16 |
| Median accelerated branches per locus | 1 |
4. Accelerated lineages in most loci under full model (M2)
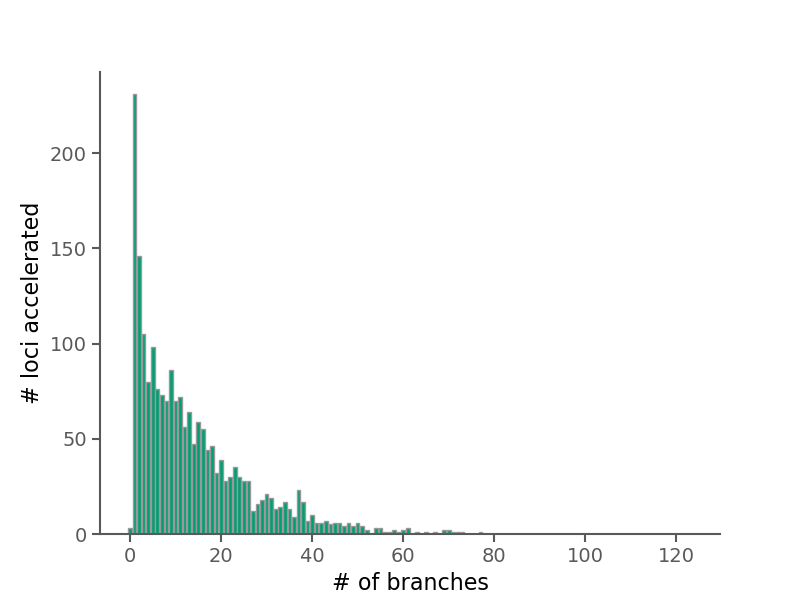
| Avg. times a branch is accelerated | 227 |
| Median times a branch is accelerated | 175 |
The 10 lineages accelerated in the most loci.